You Are What You Eat (and What You Excrete): Unveiling Diet Diversity with FoodSeq.
By Breanna Metras, PhD RDN
We know that diet is extremely impactful on an individual's microbiome, yet there is no gold standard to capture exactly what a person eats. This gap has left researchers dependent on imperfect methods to estimate dietary intake, which rely on the accuracy and honesty of participants!
Researchers have many methods of recording dietary history such as Food Frequency Questionnaires (FFQs), 24-hour recalls, dietary records, or personal food diaries. Unfortunately, there is plenty of room for human error stemming from people forgetting what they ate, not knowing how much of a food they ate, a potential lack of honesty when reporting foods, or the recall system could not capture that specific food item or ingredient. While providing good insight into diet, these strategies don’t get a perfect picture of someone's diet.
The research done thus far in my postdoc with Dr. Abigail Johnson in the Division of Epidemiology, School of Public Health at the University of Minnesota has largely focused on using bioinformatics to understand the relationship between in-depth dietary records and microbiome samples from people. We are looking at new ways to improve the quality of dietary data by bolstering it with multiple methods typically used on their own (i.e. Nutrition Data System for Research (NDSR) data and scores, MRI, VeggieMeter, BMI, FoodSeq, 16s rRNA microbiome samples).
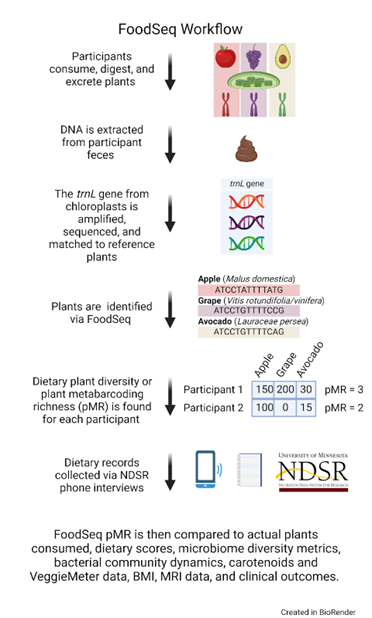
Recently, a tool commonly used for wildlife research has been adapted to help address this problem in human microbiome studies! Ecological research studying wildlife, fossils, and permafrost samples use DNA metabarcoding to sequence an animal's feces or stomach contents to better understand the foods inside; specifically for understanding prey-predator relationships and ecological networks. Genomic regions called “molecular barcodes” that identify a food species by its DNA sequence can be amplified and sequenced from the residual pool of food-derived DNA in fecal samples. If stool is already collected to understand the bacterial communities of the gut microbiome, why not also look at the plant DNA within these samples to better understand the human diet?
DNA barcoding is used to tag and amplify specific genes using conserved primers within that sample. Many microbiome researchers are familiar with barcoding using the 16S rRNA gene to identify bacterial species in a sample. Since most plants contain chloroplasts, the chloroplast gene trnL (UAA) intron and a shorter fragment of this intron (the P6 loop) have been identified as reliable primers with a robust application system for metabarcoding (1). While it may not be the perfect barcode due to low resolution of species from GenBank, it is standardized among plants and is short enough to enable amplification of degraded DNA. This is particularly helpful when considering the modern human diet which consists of many ingredients that are processed or cooked.
Using the trnL P6-loop, Dr. Lawrence David’s lab at Duke University has constructed a database to identify up to 468 commonly eaten species of plants in a person’s stool (2). To date, this technology has been labelled as “FoodSeq” and used to identify human diet composition in only a handful of studies (2-4). These studies have explored the efficacy of this technology in controlled feeding and free-eating environments in both adults and children. Researchers from the David Lab were able to compare the number of plants detected in a person's FoodSeq results with the plants captured in dietary recall. In one controlled eating study, researchers found that 38 plant taxa (79%) appeared in both the sequencing and dietary data sets, whereas only one taxon (2%) was solely recorded in the diet diaries (3), in another controlled feeding study, over 96% of plant taxa consumed were captured by FoodSeq (2).
As a diet diversity measure, the number of plants identified in a stool sample can be averaged to find the plant metabarcoding richness (pMR) (2). pMR is a marker of plant diversity that can be correlated with other dietary measures such as the HEI (Healthy Eating Index), FVS (Food Variety Score), or hPDI/uPDI/PDI (healthy, unhealthy, and overall plant-based dietary indices). Researchers have also found correlations between lifestyle factors such as age, BMI, and income with pMR (2). We often hear that consuming a diet of many plants can lead to better health outcomes, and now we may be able to put that to the test.
An interesting finding from the controlled feeding trial was that the grams of each food consumed were correlated with the number of amplicon sequence variants (ASVs) of that plant detected; indicating that the amount of a particular plant consumed could be estimated! When paired with comprehensive food recall/diaries, this method can complement dietary recall and capture foods not reported.
Although promising, the trnL P6 loop barcode isn’t able to tell the difference between plant species. This means that a person could consume brussel sprouts, cabbage, kohlrabi, or broccoli, all a part of the Brassica Oleracea species, and FoodSeq cannot tell them apart (3). Specific primers for different plant species of interest will be needed, similar to differentiating between bacterial species and strains. Additionally, coffee was not detected in any of the studies, likely due to the fact most of the plant matter is removed at the point of consumption.
Humans are omnivores, so only measuring plant matter in a diet doesn’t give us the whole picture. Researchers are working on metabarcodes to capture vertebrates consumed by people and have had moderate success in detecting different animals consumed (3,4)! One study used the stomach contents from autopsy donors to detect plants and animals consumed before the time of death. There are drawbacks with specific animal consumption as well such as differentiating between chicken and eggs, or dairy products and beef, or domesticated pork from wild boar.
FoodSeq can be used on human stool, and it can be used to check for the authenticity of ingredients, the amount of different plants used in highly processed food, or potential adulterations in foods. A study used FoodSeq on 6 processed commercial foods to evaluate the efficacy of the technology on heavily processed foods and to compare label ingredient claims with detectable plant matter (5). The research group found that this technology was able to detect highly processed and degraded plant DNA in items such as curry, a vegetable stock cube, and a herbal supplement. Surprisingly, the herbal supplement was found to be missing all ingredients listed besides dandelion (Taraxacum officinale, 99.2%) and the vegetable stock had 5 ingredients not listed, with fennel not on the label but making up 43% of the product. Thankfully, the saffron sequence contained only saffron (Crocus sativus)!
While FoodSeq is not a perfect method of identifying all plants a person has consumed, it can bolster dietary findings from other recall methods and capture unreported foods. In my research, I am starting to use this method alongside a validated telephone recall method, NDSR, to evaluate the efficacy of this technology. So far, we have found some promising results, including detecting potentially unreported foods such as cannabis edibles and wine. Perhaps one day, we will reliably identify both bacteria and foods in your gut from a single sample!
References
1. Taberlet P, Coissac E, Pompanon F, et al. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007;35(3):e14-e14. doi:10.1093/nar/gkl938
2. Petrone BL, Aqeel A, Jiang S, et al. Diversity of plant DNA in stool is linked to dietary quality, age, and household income. Proc Natl Acad Sci. 2023;120(27):e2304441120. doi:10.1073/pnas.2304441120
3. Reese AT, Kartzinel TR, Petrone BL, Turnbaugh PJ, Pringle RM, David LA. Using DNA Metabarcoding To Evaluate the Plant Component of Human Diets: a Proof of Concept. Segata N, ed. mSystems. 2019;4(5):e00458-19. doi:10.1128/mSystems.00458-19
4. Schneider J, Mas-Carrió E, Jan C, et al. Comprehensive coverage of human last meal components revealed by a forensic DNA metabarcoding approach. Sci Rep. 2021;11(1):8876. doi:10.1038/s41598-021-88418-x
5. Bruno A, Sandionigi A, Agostinetto G, et al. Food Tracking Perspective: DNA Metabarcoding to Identify Plant Composition in Complex and Processed Food Products. Genes. 2019;10(3):248. doi:10.3390/genes10030248