A game-changer for microbial isolation workflows and accelerating probiotic discovery
By Brendan Daisley, PhD (University of Guelph)
If you’ve ever worked with Sanger sequencing data, you know it can be a long and frustrating road from raw sequence files to a finalized strain library. As a microbiologist studying probiotics, I have faced the same struggles—hours spent trimming sequences, running taxonomic searches, and trying to organize data into something meaningful. That’s why we created isolateR, an R package designed to take the pain out of microbial workflows and help you focus on what really matters: discovering exciting new microbes (1).
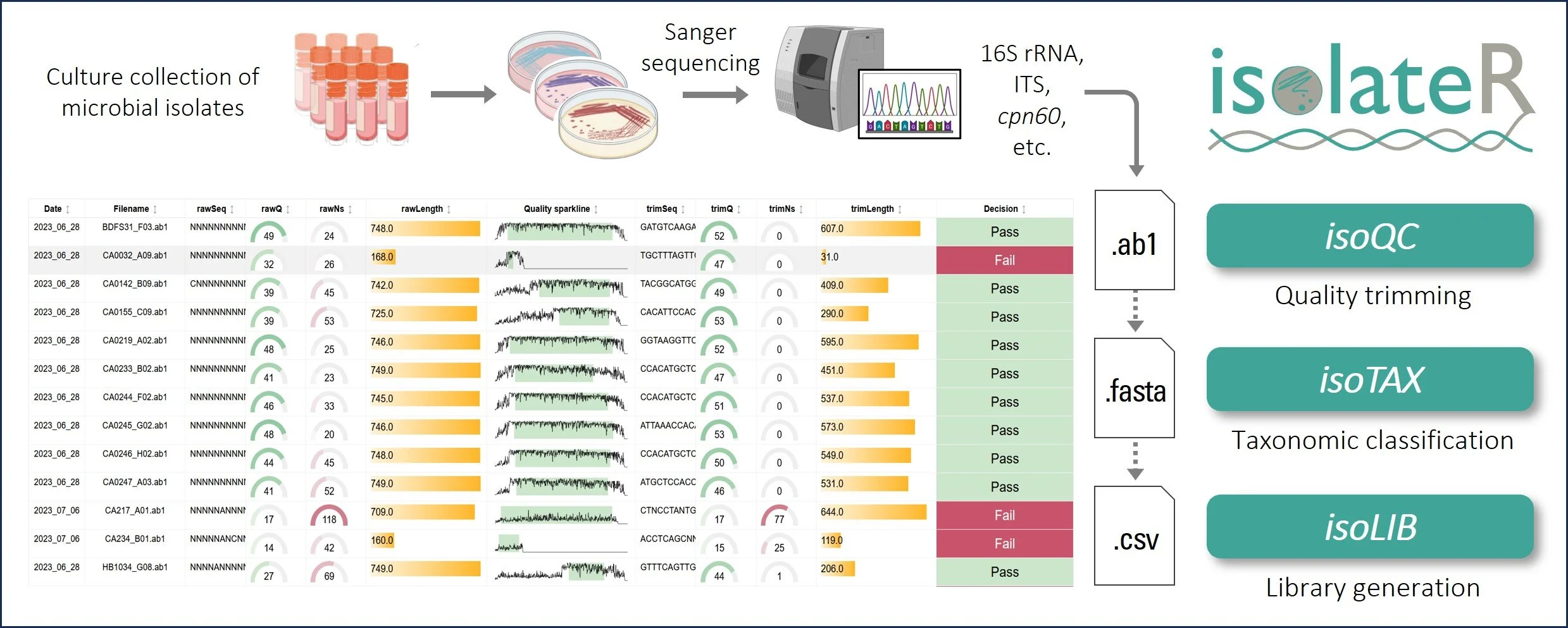
With isolateR, you can process your data, identify strains, and build libraries in just a few clicks. Here’s how it works—and how it can supercharge your microbial isolation workflows.
Step 1: Streamlined trimming and QC of sequences
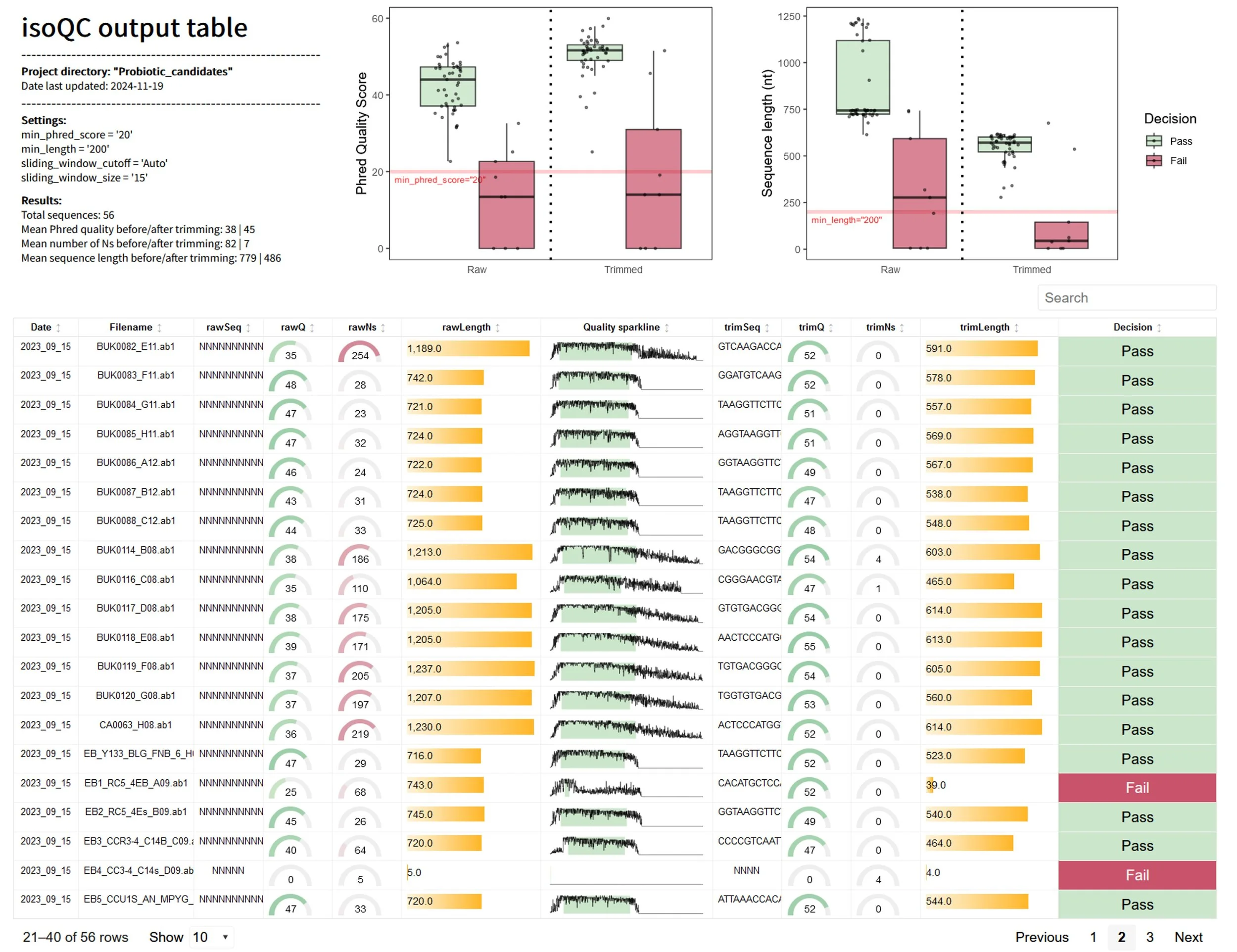
The first step in working with sequence data is cleaning it up—trimming low-quality regions, flagging ambiguous bases, and making sure only the best data moves forward. With isoQC, isolateR handles all of this automatically. You just point the tool to your .ab1 files, and isoQC processes them in batches, generating clean, trimmed sequences and interactive summaries so you can quickly see what passed and what didn’t. When I was isolating bacteria from fecal samples, isoQC saved me hours of manual trimming and quality checks. It meant I could spend more time asking questions about my data and less time wrestling with it. If you’re looking for your next probiotic superstar, this step ensures you start with high-quality candidates.
Step 2: Accurate and up-to-date taxonomic classification with isoTAX
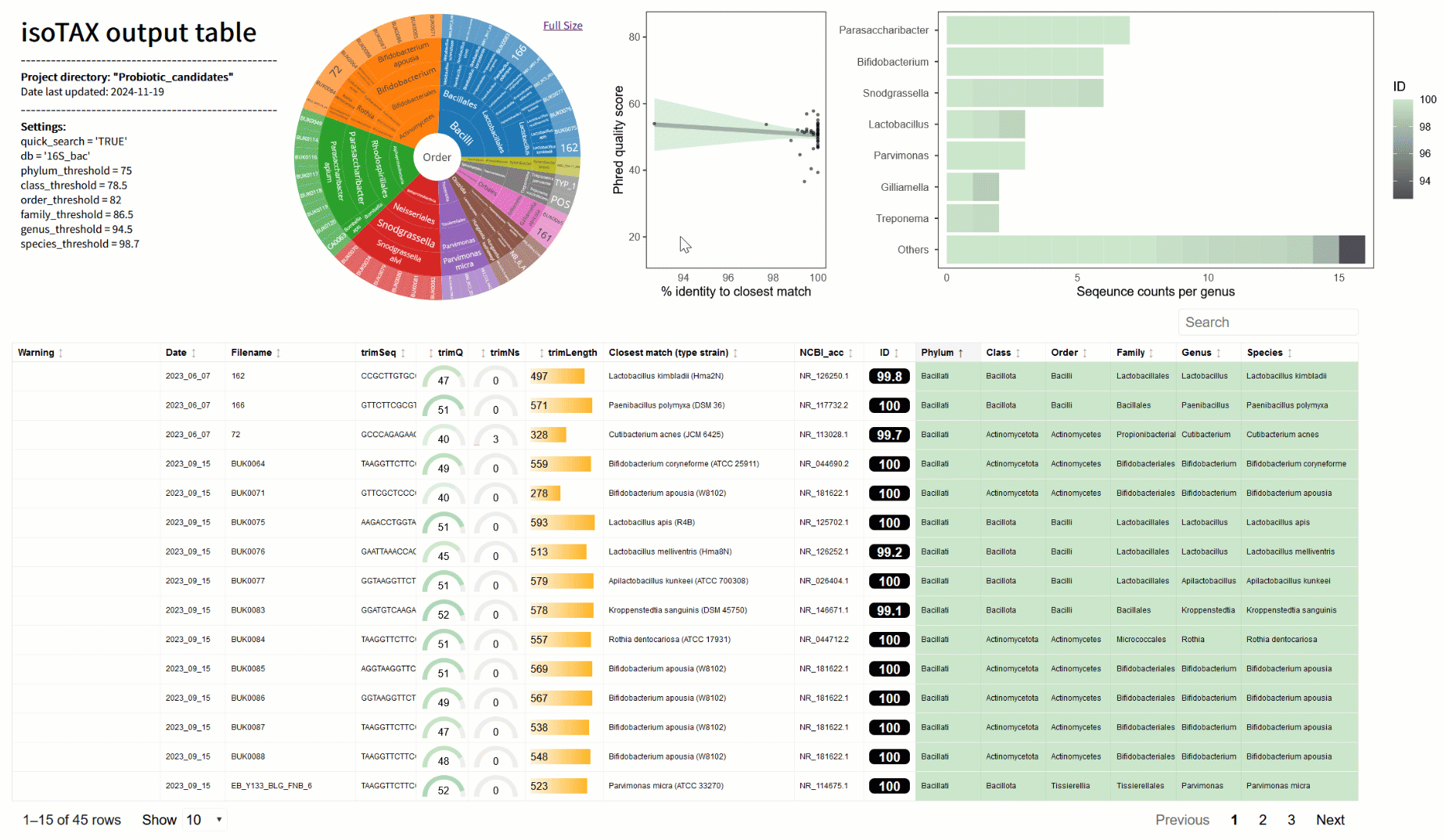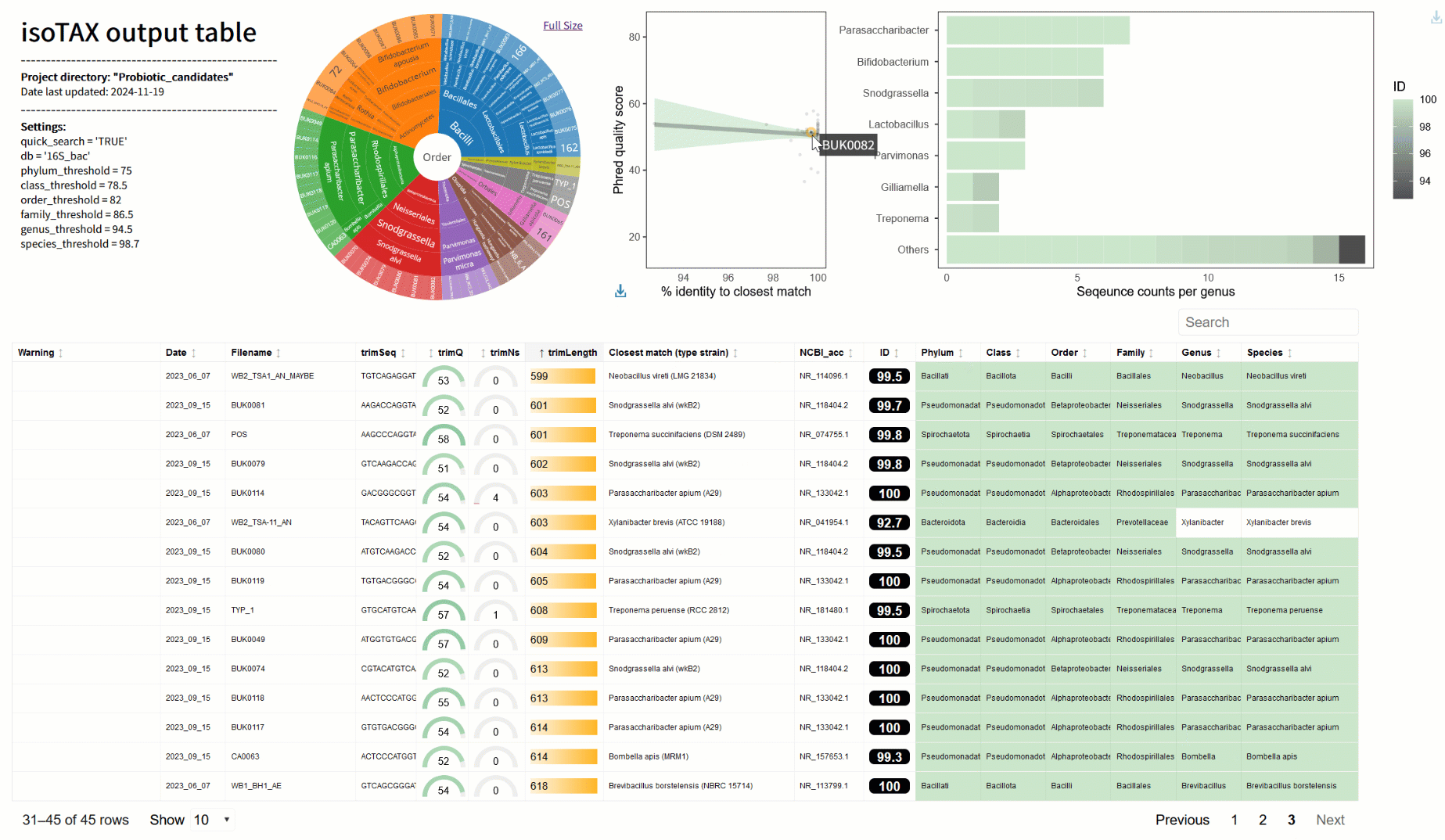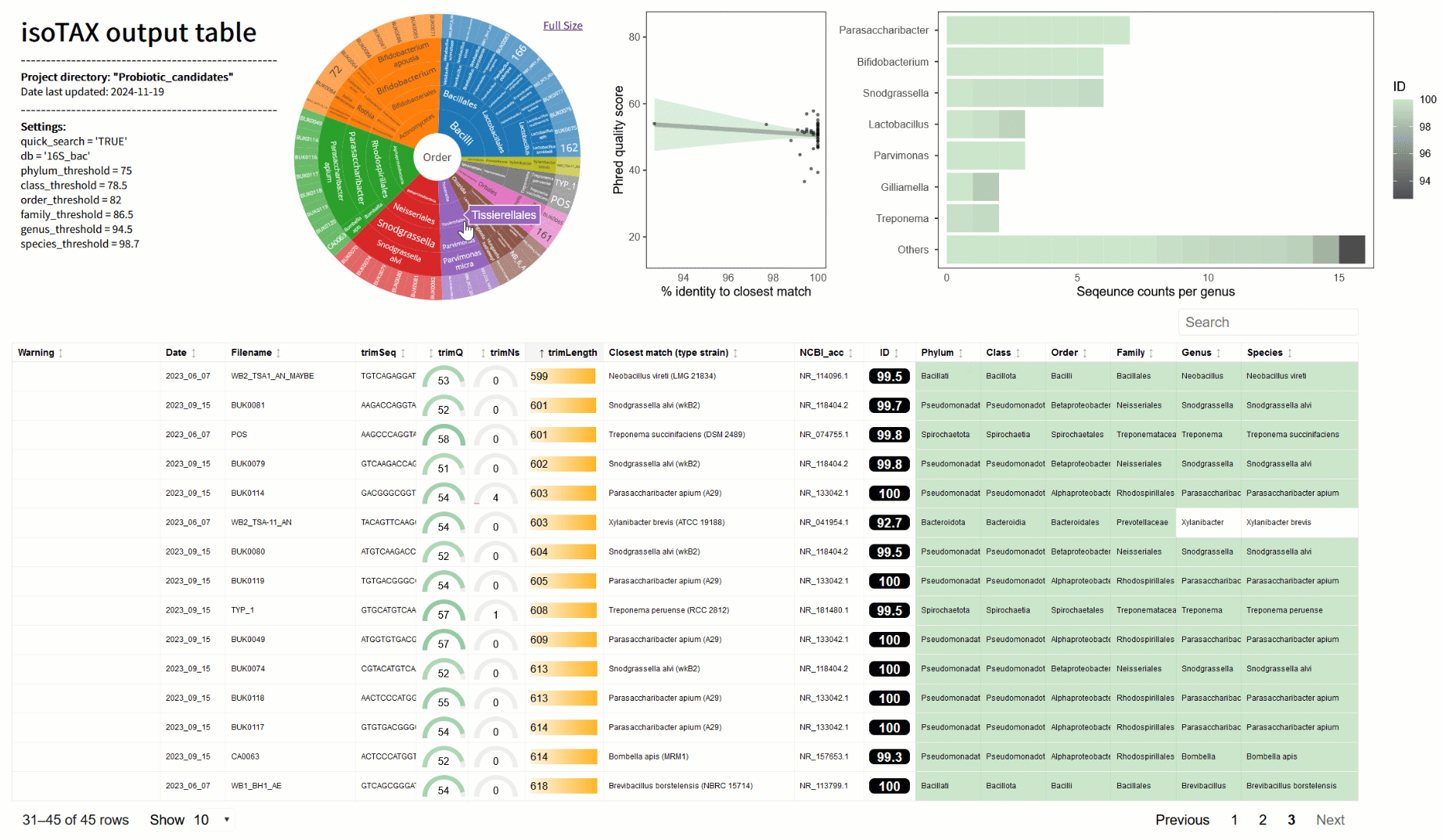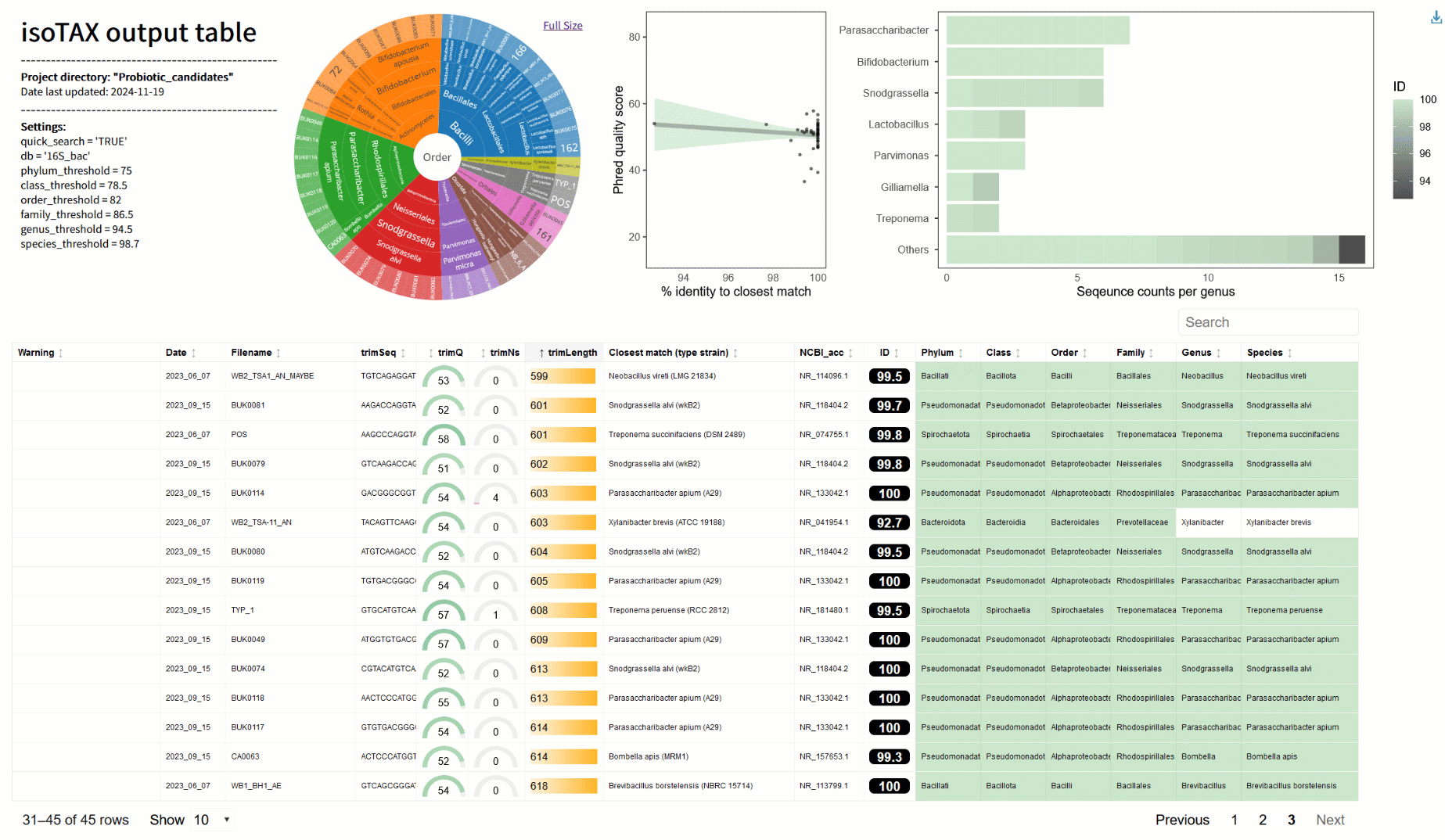
One of the biggest frustrations I’ve faced is figuring out what my sequences actually represent. BLAST is great for a quick look, but it often matches to incomplete or misclassified data (2). With isoTAX, isolateR takes a different approach, using precise global alignment to type strain databases. This means your results are more accurate and reliable—and they comply with the latest international nomenclature standards (e.g., LPSN (3)). Even better, isoTAX works with more than just 16S rRNA. Whether you’re studying fungi (with ITS) or chasing down higher resolution for bacteria (e.g., using cpn60 (4)), you can tailor the tool to your research. isolateR can also help in probiotic research, for example you can quickly identify strains from gut samples or fermented foods, flag potentially novel species, and get right to the exciting work of understanding their mechanisms in the lab.
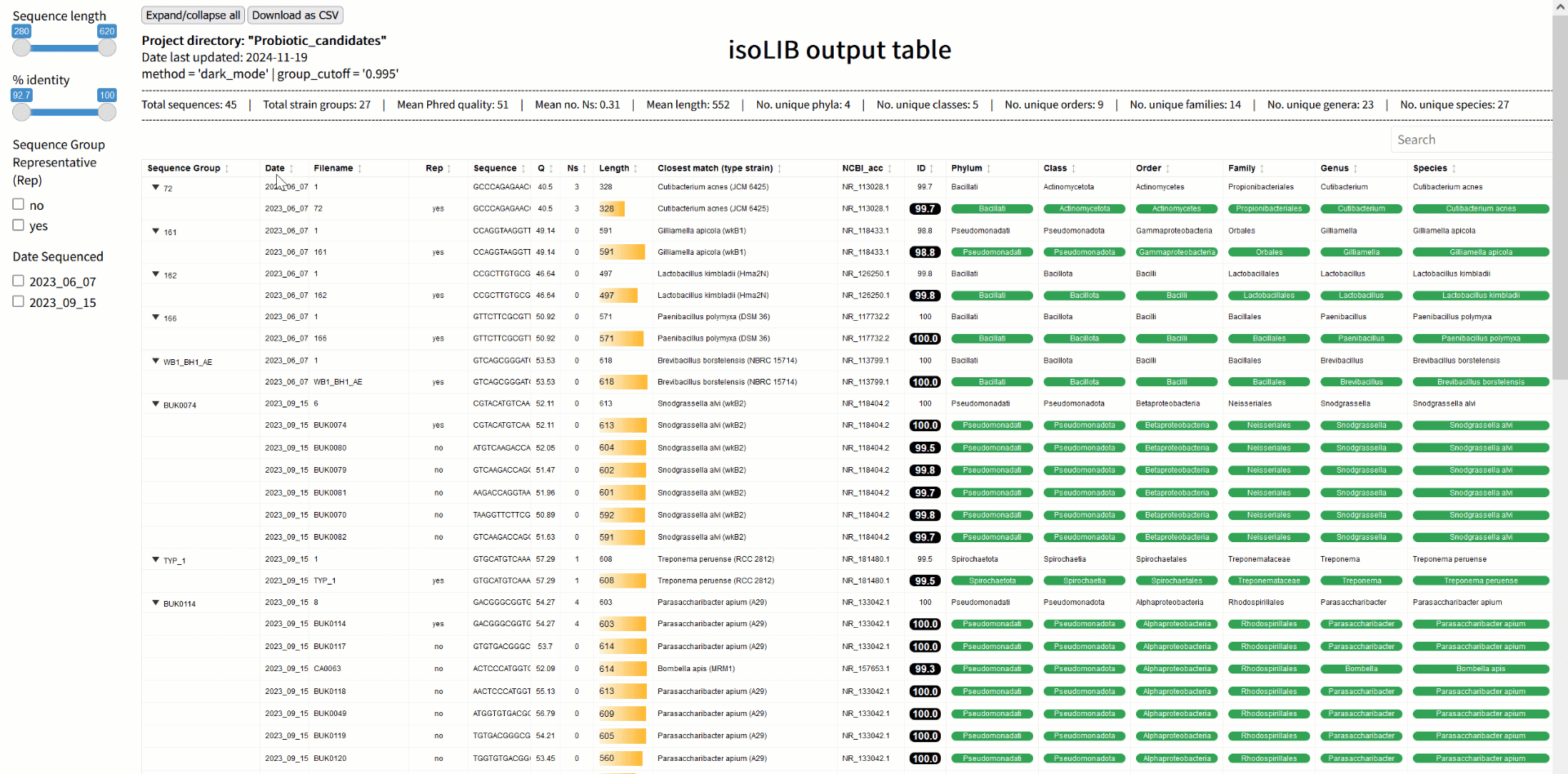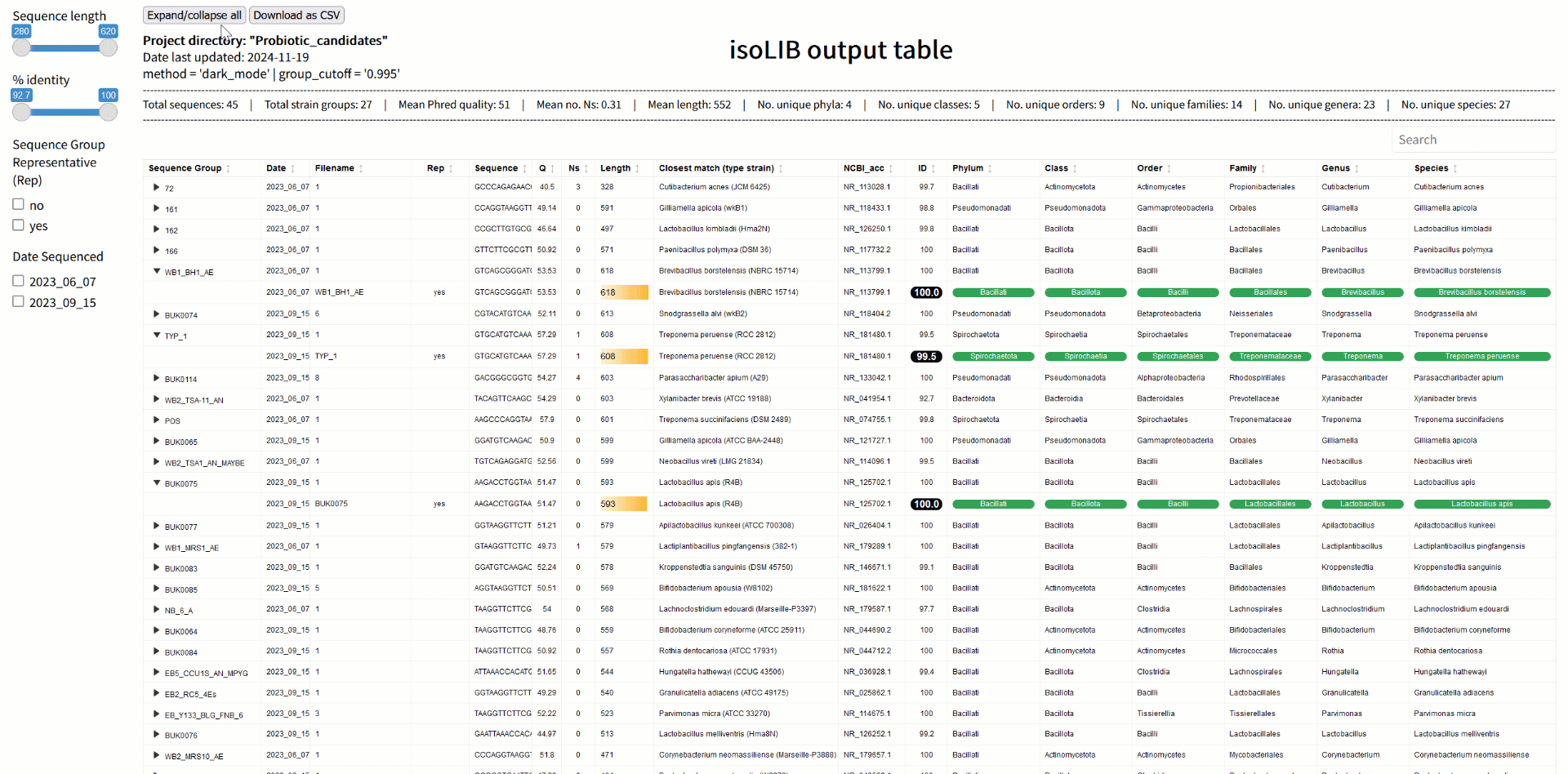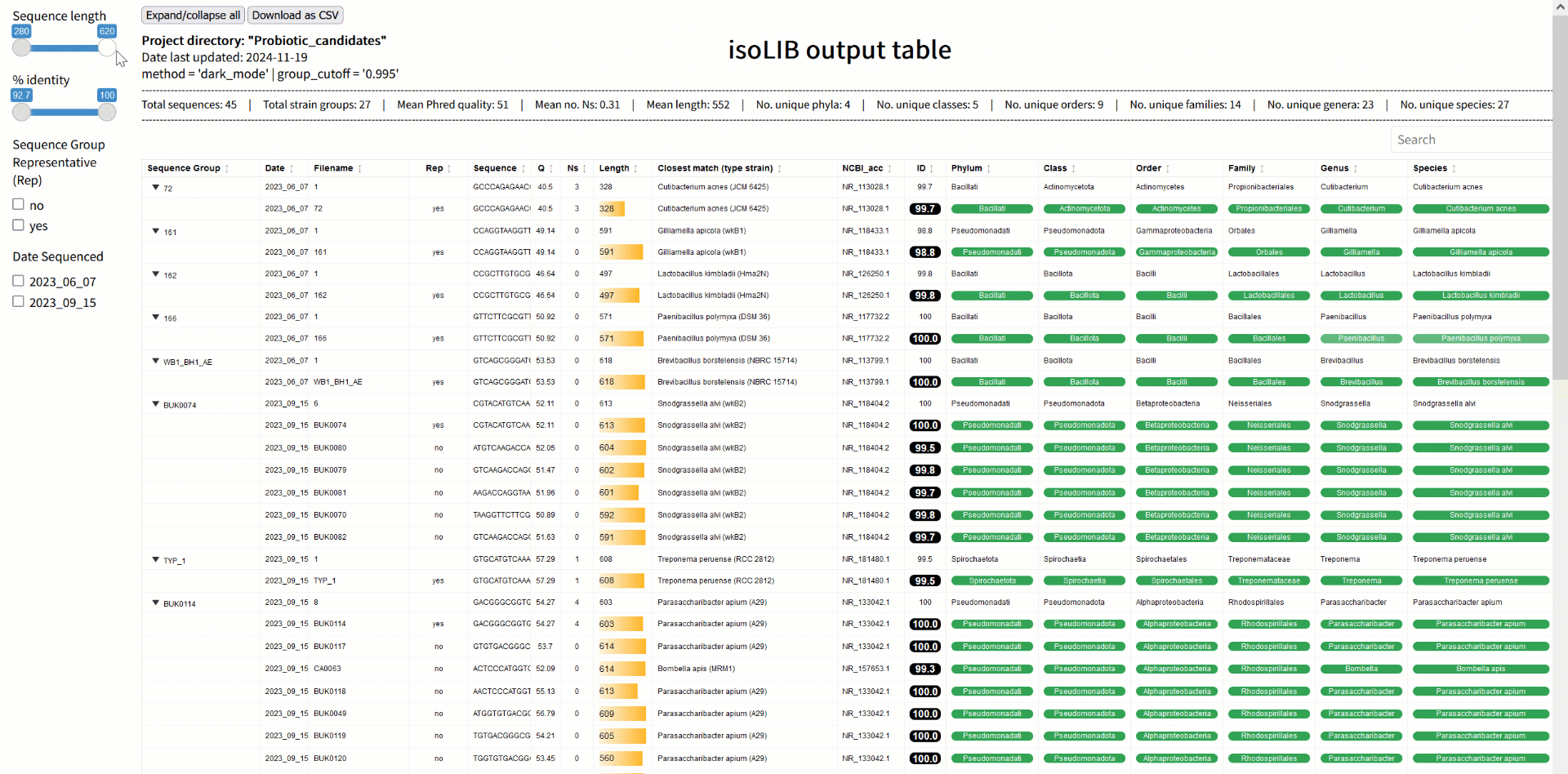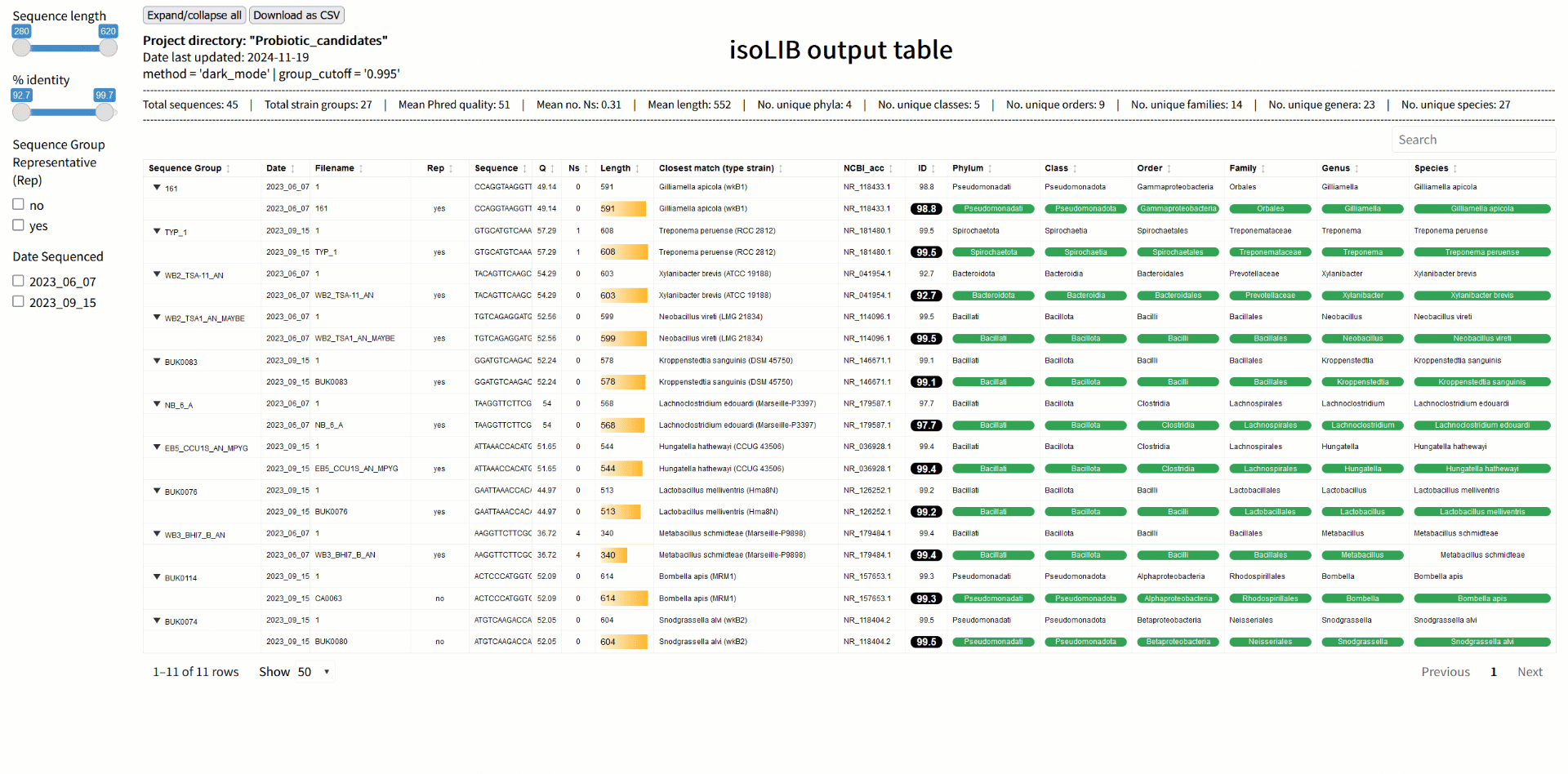
Step 3: Strain library creation with isoLIB
Once sequences are processed and classified, the next challenge is organizing them into a meaningful library. The isoLIB function simplifies this by clustering related isolates into groups based on customizable thresholds of sequence similarity. This process eliminates redundant entries, highlights unique strains, and produces an interactive, searchable HTML library. What sets isoLIB apart is its ability to integrate new isolates into existing libraries, enabling researchers to build dynamic, evolving collections. For those studying probiotics, isoLIB is a powerful tool to document strain-level diversity, pinpoint novel candidates, and compare isolates across different environments or conditions. Whether you're screening microbes from the gut, soil, or fermented foods, isoLIB provides an elegant solution for managing your findings.
A Unified Workflow for the Discovery of Novel Microbes
The best part of isolateR is how it simplifies everything. The entire workflow—from raw data to strain library—can be done in just three lines of R code. What used to take days now takes minutes, giving you more time to focus on the science and the story behind your isolates. If you’re passionate about microbes, isolateR can help you work smarter, not harder. It’s designed to make your life easier, whether you’re profiling the microbiota of fermented foods, the gut microbiota, or anything in between. By automating tedious steps and giving you tools to dig deeper into microbial diversity, isolateR turns the process of isolation into a gateway for discovery.
How To Get Started
Ultimately, isolateR was designed to address common challenges in microbial research, and it’s now freely available for everyone to use. Whether you’re a seasoned microbiologist or just starting to explore the microbial world, isolateR can help you uncover new possibilities. You can explore the package, its documentation, and examples on GitHub: https://github.com/bdaisley/isolateR.
The next big probiotic breakthrough might be sitting in your lab—it’s time to find it!
References
1. Daisley, B. et al. isolateR: an R package for generating microbial libraries from Sanger sequencing data. Bioinformatics 40, btae448 (2024).
2. Tindall, B. J., Rosselló-Móra, R., Busse, H.-J., Ludwig, W. & Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 60, 249–266 (2010).
3. Parte, A. C. LPSN—list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 42, D613–D616 (2014).
4. Vancuren, S. J. & Hill, J. E. Update on cpnDB: a reference database of chaperonin sequences. Database 2019, baz033 (2019).